|
Bioinformatics Servers (26 Servers)
|

|
ATPdock
Category: [Protein-ATP docking]
Description: A template-based method for ATP-specific protein-ligand docking method.
Journal: Bioinformatics
DOI:
10.1093/bioinformatics/btab667
Year: 2021
|

|
DeepATPseq
Category: [Protein-ATP Interaction Site Prediction]
Description: A Sequence-based ATP-Specific Binding Residue Predictor.
Journal: Analytical Biochemistry
DOI:
10.1016/j.ab.2021.114241
Year: 2021
|
|
|
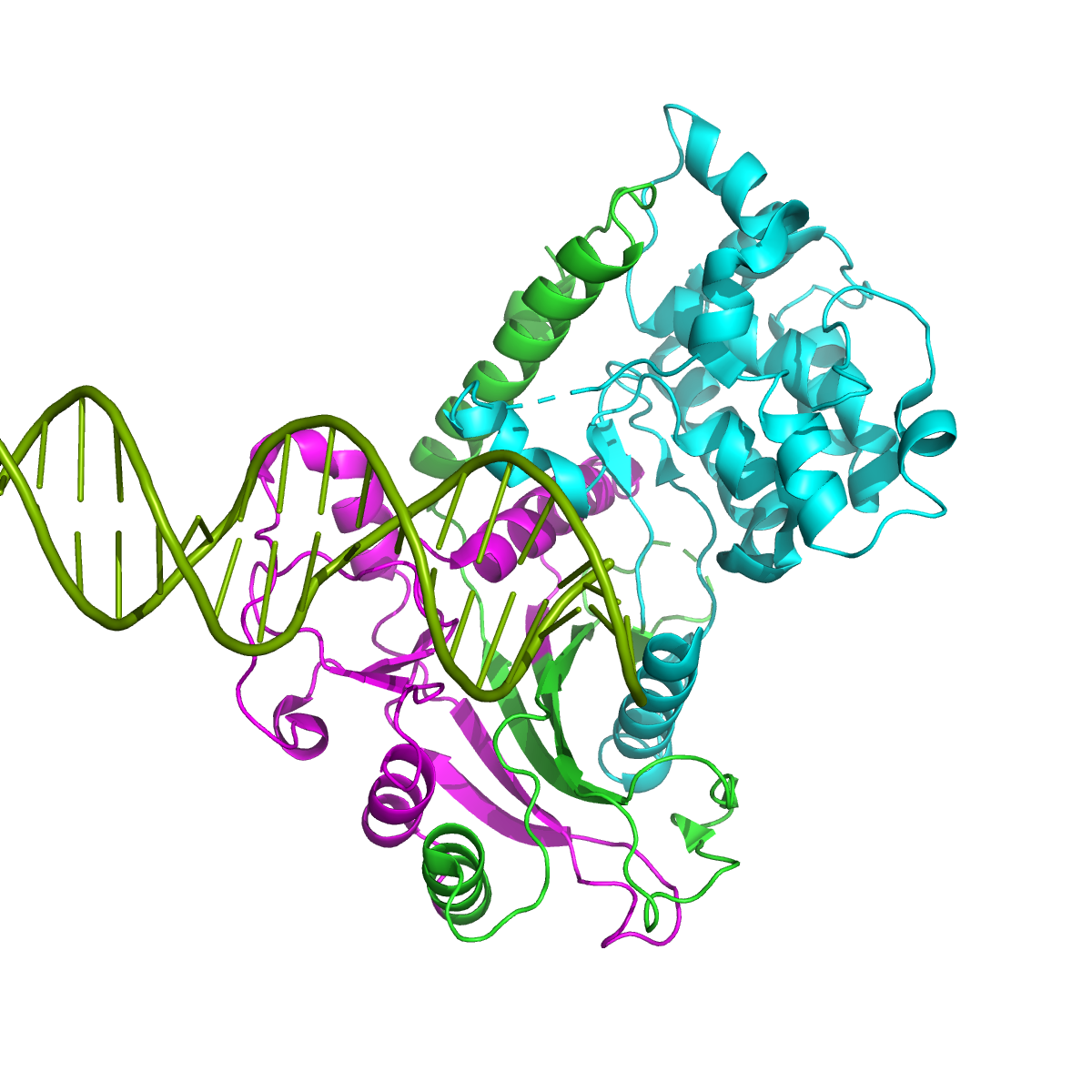
PredDBR
Category: [Protein-DNA Interaction Site Prediction]
Description: Protein-DNA Binding Residue Prediction via Bagging Strategy and Sequence-based Cube-Format Feature.
Journal: IEEE/ACM Transactions on Computational Biology and Bioinformatics
DOI:
10.1109/TCBB.2021.3123828
|

|
TargetDBP+
Category: [Protein-DNA Interaction Prediction]
Description: Enhancing the Performance of Identifying DNA-Binding Proteins via Weightedly Convolutional Features.
Journal: Journal of Chemical Information and Modeling
DOI:
10.1021/acs.jcim.0c00735
Year: 2021
|
|
|
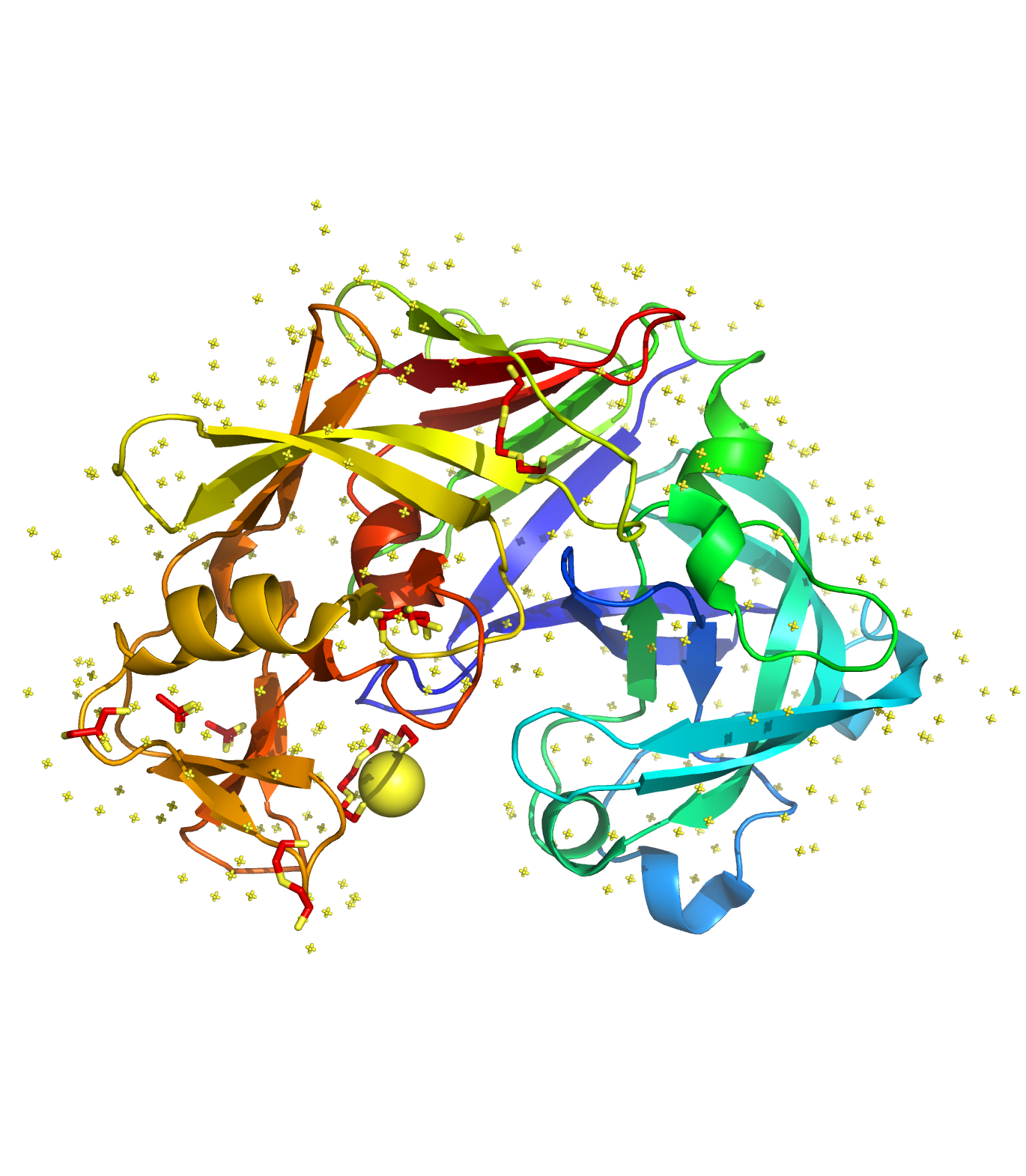
I-LBR
Category: [Ligand-Binding Residue Identification]
Description: A Query-Specific Ligand-Binding Residue Identification based on protein sequence and support vector machine.
Journal: Analytical Biochemistry
DOI:
10.1016/j.ab.2020.113799
Year: 2020
|
|
scTPA
Category: [Single-cell transcriptome analysis]
Description: A web tool for single-cell transcriptome analysis of pathway activation signatures.
Journal: Bioinformatics
DOI:
10.1093/bioinformatics/btaa532
Year: 2020
|
|
|
ResPRE
Category: [Protein residue-residue contact-map prediction]
Description: ResPRE is an algorithm for protein residue-residue contact-map prediction.
Journal: Bioinformatics
DOI: 10.1093/bioinformatics/btz291
Year: 2019
|

|
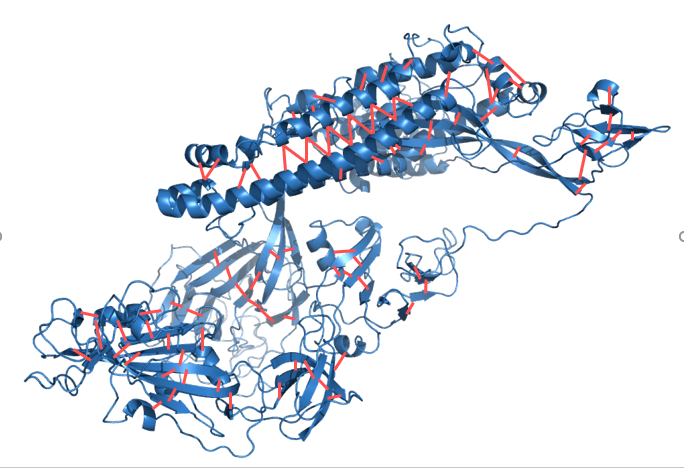
DEMO
Category: [Automated Assembly of multi-domain protein]
Description: DEMO (Domain Enhanced MOdeling) is a method for automated assembly of full-length structural models of multi-domain proteins.
Journal: Proceedings of the National Academy of Sciences of the United States of America
DOI: 10.1073/pnas.1905068116
Year: 2019
|
|
|
PoCom
Category: [Generating nor-order structural comparsion/alignment of two protein pockets]
Description: PoCom is an algorithm designed for quckly and accurately generating non-order structural comparison/alignment of two protein pockets.
|
|
PPS-search
Category: [Automatically searching protein pockets]
Description: PPS-search is a server for automatically searching protein pockets with similar structures.
|
|
|
TargetDBP
Category: [DNA-Binding Proteins]
Description: Accurate DNA-Binding Protein Prediction via Sequence-based Multi-View Feature Learning.
Journal: IEEE/ACM Transactions on Computational Biology and Bioinformatics
DOI: 10.1109/TCBB.2019.2893634
Year: 2019
|

|
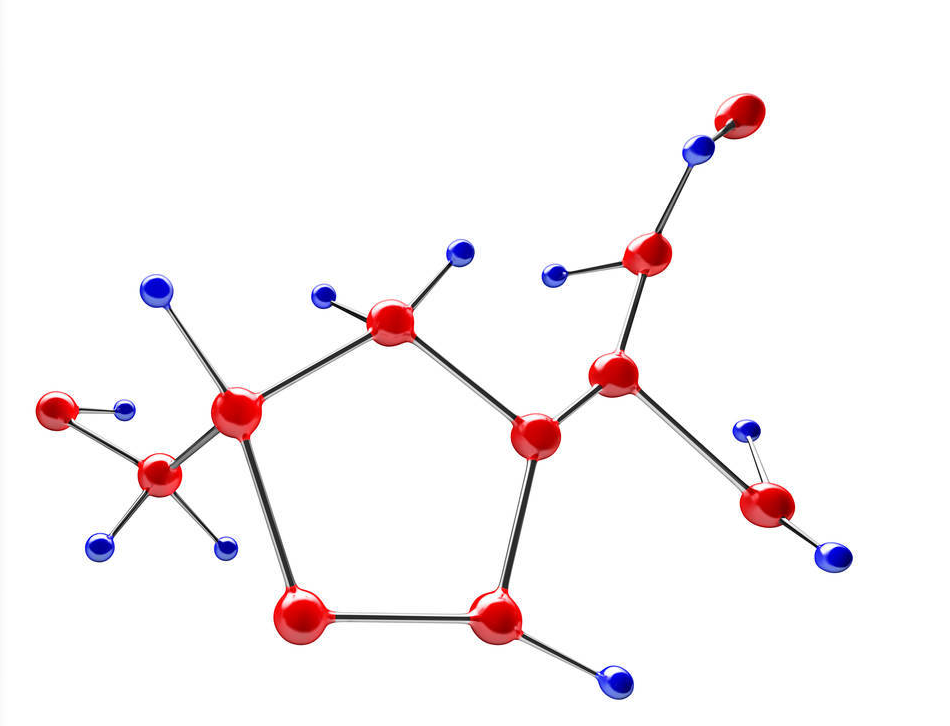
LS-align
Category: [Ligand structural alignment]
Description: LS-align is an algorithm designed for atom-level structural comparison of ligand molecules.
Journal: Bioinformatics
DOI: 10.1093/bioinformatics/bty081
Year: 2018
|
|

|
ATPbind
Category: [Protein-ATP Binding sites]
Description: Accurate protein-ATP binding site prediction by combining sequence-profiling and structure-based comparisons.
Journal: Journal of Chemical Information and Modeling
DOI: 10.1021/acs.jcim.7b00397
Year: 2018
|

|
TargetDNA
Category: [Protein-DNA binding sites]
Description: Protein-DNA Binding Residues by Weightedly Combining Sequence-based Features and Boosting Multiple SVMs.
Journal: IEEE/ACM Transactions on Computational Biology and Bioinformatics
DOI: 10.1109/TCBB.2016.2616469
Year: 2017
|
|
|
TargetCrys
Category: [Protein Crystallization Prediction]
Description: Protein crystallization prediction by fusing multi-view features with two-layered SVM.
Journal: Amino Acids
DOI: 10.1007/s00726-016-2274-4
Year: 2016
|
|
TargetNUCs
Category: [Protein-Nucleotide Interaction]
Description: KNN-based Dynamic Query-Driven Sample Rescaling Strategy for Class Imbalance Learning.
Journal: Neurocomputing
DOI: 10.1016/j.neucom.2016.01.043
Year: 2016
|
|
|
TargetGDrug
Category: [Protein-Drug Interaction]
Description: Predicting GPCR-Drug Interactions by Random Forest with Drug Association Matrix Based Post-processing Procedure.
Journal: Computational Biology and Chemistry
DOI: 10.1016/j.compbiolchem.2015.11.007
Year: 2016
|
|
TargetFreeze
Category: [Antifreeze Protein Prediction]
Description: Identifying Antifreeze Proteins via a Combination of Weights using Sequence Evolutionary Information and Pseudo Amino Acid Composition.
Journal: Journal of Membrane Biology
DOI: 10.1007/s00232-015-9811-z
Year: 2015
|
|

|
OSML
Category: [Protein-Nucleotide Interaction]
Description: A Query-Driven Dynamic Machine Learning Model for Predicting Protein-Ligand Binding Sites.
Journal: IEEE Transactions on NanoBioscience
DOI: 10.1109/TNB.2015.2394328
Year: 2015
|

|
TargetDisulfide
Category: [Disulfide Connectivity Prediction]
Description: Disulfide Connectivity Prediction Based on Modelled Protein 3D Structural Information and Random Forest Regression.
Journal: IEEE/ACM Transactions on Computational Biology and Bioinformatics
DOI: 10.1109/TCBB.2014.2359451
Year: 2015
|
|
|
TargetSOS
Category: [Protein-Nucleotide Interaction]
Description: A supervised over-sampling (SOS) predictor for targeting protein-nucleotide binding residues.
Journal: PLoS ONE
DOI: 10.1371/journal.pone.0107676
Year: 2014
|

|
TargetVita
Category: [Protein-Vitamin Interaction]
Description: Enhancing Protein-Vitamin Binding Residues Prediction by Multiple Heterogeneous Subspace SVMs Ensemble.
Journal: BMC Bioinformatics
DOI: 10.1186/1471-2105-15-297
Year: 2014
|
|

|
TargetS
Category: [Protein-Ligand Interaction]
Description: A ligand-specific template-free predictor for targeting protein-ligand binding sites.
Journal: IEEE/ACM Transactions on Computational Biology and Bioinformatics
DOI: 10.1109/TCBB.2013.104
Year: 2013
|
|
COMSPA
Category: [Protein Structural Classes Prediction]
Description: Predicting Protein Structural Classes From Primary Sequence by Learning Multi-View Features in Complex Spac.
Journal: Amino Acids
DOI: 10.1007/s00726-013-1472-6
Year: 2013
|
|

|
TargetATPsite
Category: [Protein-ATP Interaction]
Description: A Template-free Method for ATP Binding Sites Prediction with Residue Evolution Image Sparse Representation and Classifier Ensemble.
Journal: Journal of Computational Chemistry
DOI: 10.1002/jcc.23219
Year: 2013
|

|
TargetATP
Category: [Protein-ATP Interaction]
Description: Improving Protein-ATP Binding Residues Prediction by Boosting SVMs with Random Under-Sampling.
Journal: Neurocomputing
DOI: 10.1016/j.neucom.2012.10.012
Year: 2013
|
|